From Vijaywada to Nagpur as a part of my lecturing...
Read MoreDr. Suparna Mitra
With a PhD in Bioinformatics and 14+ years of research experience in applying metagenomics and metatranscriptomics to medical sciences I am active in research and teaching at University of Leeds.
I teach and supervise students and researchers Medical and Applied Statistics, Bioinformatics and the emerging field of Metagenomics in the Faculty of Medicine and Heath and School of Biological Sciences.
I am also module lead for ENQUIRE2 at University of Leeds medical degree program (MBChB), where second year medical students learn how to do research and apply Statistics in Medicine.

About Me
The majority of my research at University of Leeds (UoL) involves applying metagenomics and metatranscriptomics to medical sciences, especially the complex gut microbiome which is a major potential reservoir for antimicrobial resistance genes that may be selected for and/or transferred to/between potential pathogens.
With an interdisciplinary research background, I am inspired to apply my knowledge and experience in developing multivariate methods for ’omics data analyses that can identify potential applications in the real-world. During my research career, I have >14 years of experience in working both theoretical (statistics/bioinformatics) and applied (biostatistics/ medical/ biology/ infection control) research groups and various projects. Whilst still in the mid stage of my career my published research( >40 article and book chapters) has a good impact: total >5000 citations (H-index=21).
For past few years I have focused my research to understand the impact of Unrestricted Antibiotic Usage on the Gut Microbiome and Antibiotic Resistance Genes (ARGs) among relatively healthy population. Our pilot study in Bangladesh is the first of its kind. There is no other study trying to investigate the unrestricted use of antibiotics in generally healthy population and exploring other possible sources of AMR, like animal and environment.
Mirroring this study in India will give us the comparative factor in neighbouring country. Multiple locations will enable us to include effect of various culture and food habit. Further my plan is to extend this as a prospective cohort study, so that we can investigate the changes of microbiome within individuals’ and associate them with different groups of antibiotics to identify key, patho-physiologically/functionality relevant microbiome changes (taxonomy and functions).
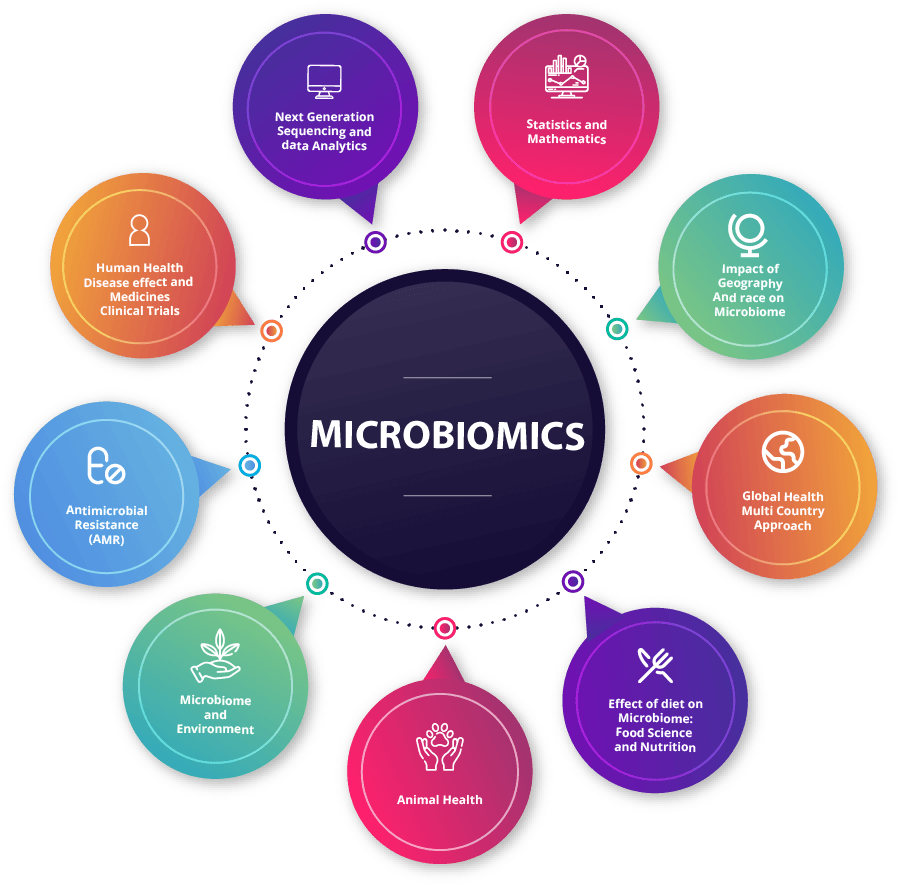
Key Areas of Research Interest
- Metagenomics.
- Metatranscriptomics.
- Whole Genome Shotgun Sequencing.
- Application in Medical Studies.
- Metagenome Assembly.
- Application in Personalised Medicine.
- Taxonomic and Functional annotation.
- Clinical Trials.
- Applied Statistics & Algorithms.
- Sequencing Data Analysis.
- Health Care Associated Infection.


Our Microbiome: Good or Bad ?
The human body is home to several millions microbes which is called microorganisms all together . Some microbes make us sick, others are important for our health. The most common types are bacteria, viruses and fungi. They can be both helpful and potentially harmful. Most are symbiotic (where both the human body and microbiota benefit) and some, in smaller numbers, are pathogenic (promoting disease). In a healthy body, pathogenic and symbiotic microbiota coexist without problems. We want our body in a equilibrium. And nurture our good warriors with good food and healthy lifestyle. But often with unhealthy diet, and bad lifestyle, and unrestricted use of antimicrobials we ruin our healthy gut flora.
My work is focus on this How we can study and understand our microbiome within our body and in the environment. How we can restrict the use of antimicrobials and save the life saving drugs for future generations. I want to be part of a shared platform for tackling global AMR challenge by connecting interdisciplinary research communities and addressing recent demands in medical science combining methodologies for data analyses with advanced sequencing techniques.
Good Bacterial Flora

Bifidobacteria
Escherichia coli

Lactobacilli
Nurture them or ruin them?
Bad Bacterial Flora

Campylobacter
Enterococcus Faecalis

Clostridium Difficile
Antibiotics and Alternative Strategies for Combating Antimicrobial Resistance Conference
Having an incredible time at the "Antibiotics and Alternative Strategies for Combating Antimicrobial Resistance Conference" at Coimbatore arranged by Sri Krishna Arts and Science College, Coimbatore India as an invited speaker. Thanks to Academy of Medical Science for supporting my travel to India. Also thanks to the organisers for inviting me and arranging my stay here at Coimbatore
Michael Beverley Innovation Fellowship Interview - Suparna Mitra
June 2023
December was full of excitement. Most of my research involves the complex gut microbiome which is a major potential reservoir for antimicrobial resistance (AMR) genes. Major problem in Low- and Middle-income Countries
(LMICs is) the availability of antibiotics in local pharmacies and unrestricted use in generally healthy populations.
I had a wonderful trip in Dhaka, Bangladesh. Great achievement of our collaborators, our AMR
Microbiome project is Dhaka (#DhakagutAMR) is moving in full swing. I have visited Dhaka and we had a community engagement day and lots of interest from the participants. . All thanks to my Michael Beverley fellowship #MBIF.
Our pilot study
“Impact of Unrestricted Antibiotic Usage on the Gut Microbiome and Antibiotic Resistance Genes (ARGs) among Bangladeshi Population”.

Microbiome and Antimicrobial Resistance
Major Grants
2023-24 – Hamied Foundation UK-India AMR Visiting Fellowships – £6000
Principal Investigator.
Principal Investigator.
£7500. Principal Investigator
2022-2023 – Studying longitudinal microbiome shift and antimicrobial resistance (AMR) in association with unrestricted antibiotic usage in a poor Community.; International Strategic Fund (ISF); Awarded value (100%): £4000.
Principal Investigator

Teaching and Student supervisions

Teaching courses
- I started teaching at the University of Leeds on ‘Research, Evaluation and Special Studies (RESS) 1-Human Biomarker Project and ‘RESS Special Studies Project (SSP)’ (MEDI1213) from 2018. Due to unique and up-to-date topics in research for SSP projects, my topics were in high demand and that lead me to continue serving in RESS-SSP for past four years.
- Since 2021 I am acting as MBChB RESS2 co-lead and for 2023-24 solely lead the module transfer from RESS2 to ENQUIRE2 (MEDI2224).
- Redesigned and taught all lectures and practical labs in ‘Analytical Skills in Precision Medicine’ module (BIOL5322M) since 2020.
- Designed and delivered next generation sequencing (NGS) tutorial on ‘Molecular Biology of Cancer’ module (MEDM5141M) since 2020.
- Delivered two NGS lecture on MSc Cancer biology and molecular oncology module (MEDM5221) from Jan 2021. and supervised Literature research projects for several students over past three years.
Student Experience
I am passionate about teaching and spending time with my students. I have obtained 8 years of teaching experience and I have always focus on Students’ experience and well-being. I am glad to receive often very good feedback like:
- “Never met a teacher more enthusiastic before! grateful for the insight given into real challenges in research”
- “My tutor was brilliant and responded to emails promptly. She was always happy to give feedback”
- “It’s great-I love how thorough it is, much appreciated”, “very comprehensive”
- “definitely good to have examples along the way, very straightforward, thank you”
- UAF representative of the FMH Equality and Inclusion Committee,
- Leeds Omics group
- Southeast Asia Regional Group
Publications
Journal Articles
Book Chapters
Christian Zenner, Lisa Chalklen, Helena Adjei, Matthew J Dalby, Suparna Mitra, Emma Cornwell,
Alex Shaw, Kathleen Sim, Simon J. Kroll, Lindsay J. Hall. Faecal cytokine levels of preterm infants coupled with microbiome profiles represent a potential non-invasive method to predict severity of necrotizing enterocolitis.
Gastro Hep Advances. 2023. (submitted)
Suparna Mitra, Christopher J Stewart, Andrew Nelson, James S Hampton, Andrea C Masi, Sara Manning, Christopher A Lamb, Linda Sharp, Mark A Hull, Colin J Rees. Long-term stability of the faecal microbiome profile in immunochemical test (FIT) samples.
Gut letter. 2023 (submitted)
James S Hampton, Sara Koo, Christina Dobson, Christopher J Stewart, Laura J Neilson, Kyle Montague, Suparna Mitra, John Whelpton, Caroline Addison, Phil Kelly, Stephen Rushton, Mark A Hull, Linda Sharp, Colin J Rees. The COLO-COHORT (Colorectal Cancer Cohort) study: Protocol for a multi-centre, observational research study and development of a consent-for-contact research platform.
Colorectal Dis. 2022.
https://doi.org/10.1111/codi.16160
Anthony Buckley, Ines B. Moura, Norie Arai, William Spittal, Emma Clark, Yoshihiro Nishida,Hannah C. Harris, Karen Bentley, Georgina Davis, Dapeng Wang, Suparna Mitra, Takanobu Higashiyama, Mark H. Wilcox. Trehalose-Induced Remodelling of the Human Microbiota Affects Clostridioides difficile Infection Outcome in an In Vitro Colonic Model: A Pilot
Study Front. Cell. Infect. Microbiol. 2021. https://doi.org/10.3389/fcimb.2021.670935
Gael Nana, Suparna Mitra, Henry Watson, Caroline Young, Henry M Wood, Sarah L Perry, Amanda DRace, Philip Quirke, Giles J Toogood, Paul M Loadman, Mark A Hull Luminal Bioavailability of Orally Administered Ω-3 PUFAs in the Distal Small Intestine, and Associated Changes to the Ileal Microbiome, in Humans with a Temporary Ileostomy 2021 (Under review).
The Journal of Nutrition 2021. https://doi.org/10.1093/jn/nxab113
Christopher Rooney, Kulveer Mankia, Suparna Mitra, Ines Moura, Paul Emery, Mark Wilcox,Perturbations of the gut microbiome in anti-CCP positive individuals at risk of developing rheumatoid arthritis.
Rheumatology (Oxford) 2020.
https://doi.org/10.1093/
Journal Articles
Shayantan Mukherji, Anandita Ghosh, Chandrima Bhattacharyya, Ivy Mallick, Anish Bhattacharyya,Suparna Mitra and Abhrajyoti Ghosh. Molecular and culture-based surveys of metabolically active hydrocarbon-degrading archaeal communities in Sundarban mangrove sediments.
Ecotoxicol Environ Saf. 2020. https://doi.org/10.1016/j.ecoenv.2020.110481
rheumatology/keaa792
Cristina Alcon-Giner, Matthew J Dalby, Shabhonam Caim, Jennifer Ketskemety, Alex Shaw, Kathleen Sim, Melissa A E Lawson, Raymond Kiu, Charlotte Leclaire, Lisa Chalklen, Magdalena Kujawska,Suparna Mitra, Fahmina Fardus-Reid, Gustav Belteki, Katherine McColl, Jonathan R Swann, J Simon Kroll, Paul Clarke , Lindsay J Hall. Microbiota supplementation with Bifidobacterium and Lactobacillus modifies the preterm infant gut microbiota and metabolome: an observational study Cell Rep Med. 2020. https://doi.org/10.1016/j.xcrm.2020.100077
Gourvendu Saxena§, Suparna Mitra§, Ezequiel Marzinelli, Xie Chao, Toh Jun Wei, Peter D. Steinberg,Rohan Williams, Staffan Kjelleberg, Federico Lauro and Sanjay Swarup. Metagenomics Reveals the Influence of Land Use and Rain on the Benthic Microbial Communities in a Tropical Urban Waterway. mSystems. 2018, 5;3(3). pii: e00136-17. https://doi.org/10.1128/mSystems.
Henry Watson§, Suparna Mitra§, Fiona C. Croden, Morag Taylor, Henry M. Wood, Sarah L. Perry,Jade Spencer, Philip Quirke, Giles J Toogood, Clare L Lawton, Louise Dye, Paul M Loadman, Mark A Hull. A randomised trial of the effect of omega-3 polyunsaturated fatty acid supplements on the human intestinal microbiota. Gut. 2017, 67(11). https://gut.bmj.com/content/67/11/1974
Cristina Alcon-Giner, Shabhonam Caim, Suparna Mitra, Jennifer Ketskemety, Udo Wegmann, John Wain, Gusztav Belteki, Paul Clarke, Lindsay J. Hall. Optimisation of 16s rRNA gut microbiota profiling of extremely low birth weight infants. BMC Genomics. 2017, 18(1):841.
https://doi.org/10.1186/s12864-017-4229-x.
Suparna Mitra, Caroline Chilton, Jane Freeman, Henry Wood, Phillip Quirke, Morag Taylor, Richard J. Vickers, Mark H. Wilcox. Preservation of Gut Microbiome Following Ridinilazole vs.Fidaxomicin Treatment of Clostridium difficile Infection. Abstract: Open Forum Infectious Diseases. 2017 :4 (Suppl1). (presented in ID Week Oct’17)
Book Chapters
2024 Diogo Onuselogu, Suparna Mitra How have massively parallel sequencing technologies furthered our understanding of oncogenesis and cancer progression? (Submitted in High Throughput genome techniques, Springer Nature), 2024
2023 Christopher M. Rooney, Suparna Mitra A practical guide to 16S rRNA microbiome analysis in musculoskeletal disorders in Metagenomic Data Analysis (Mitra S, ed.), 2023, Springer Nature. https://link.springer.com/protocol/10.1007/978-1-0716-3072-3_5.
2023
Shannon Jenkins, Suparna Mitra
From Microbial Genomics to Metagenomics in the era of next-generation sequencing in Metagenomic Data Analysis (Mitra S, ed.), 2023, Springer Nature. https://link.springer.com/protocol/10.1007/978-1-0716-3072-3_1


Source of my perseverance
I always truly believe Look no further, your motivation is inside yourself and is only directed to the things you love passionately! And for me I get my motivation from my loved ones: My parents, My loving husband and my amazing son. I truly love my life, my family and my work.
I am determined to prove those wrong who believes a woman with a child can not do something. rather i would say: there is no such thing that a mother can not do.



My Blogs
Lorem ipsum dolor sit amet, consectetur adipiscing elit. Ut elit tellus, luctus nec ullamcorper mattis, pulvinar dapibus leo.
Antibiotics and Alternative Strategies for Combating Antimicrobial Resistance Conference
Having an incredible time at the “Antibiotics and Alternative Strategies...
Read MoreAwarded the Hamied Foundation UK-India AMR Fellowship from the Academy of Medical Sciences
Excited to share that I’ve been awarded the Hamied Foundation...
Read More